The paper in Nature is here: https://rdcu.be/1ebq
Currently, only two types of phototrophic strategies are known to exist on Earth, chlorophyll-based (oxygenic and anoxygenic photosynthesis) and retinal-based (rhodopsin phototrophy). In an attempt to find new forms of phototrophy, we recently developed a functional metagenomics screen to enable the detection of environmental fosmid clones (in Escherichia coli) that produce proton gradients upon illumination. Using this functional screen with a marine library, we were able to find some already known proteorhodopsins, and as expected most came from Proteobacteria (Pushkarev and Béjà, 2016).

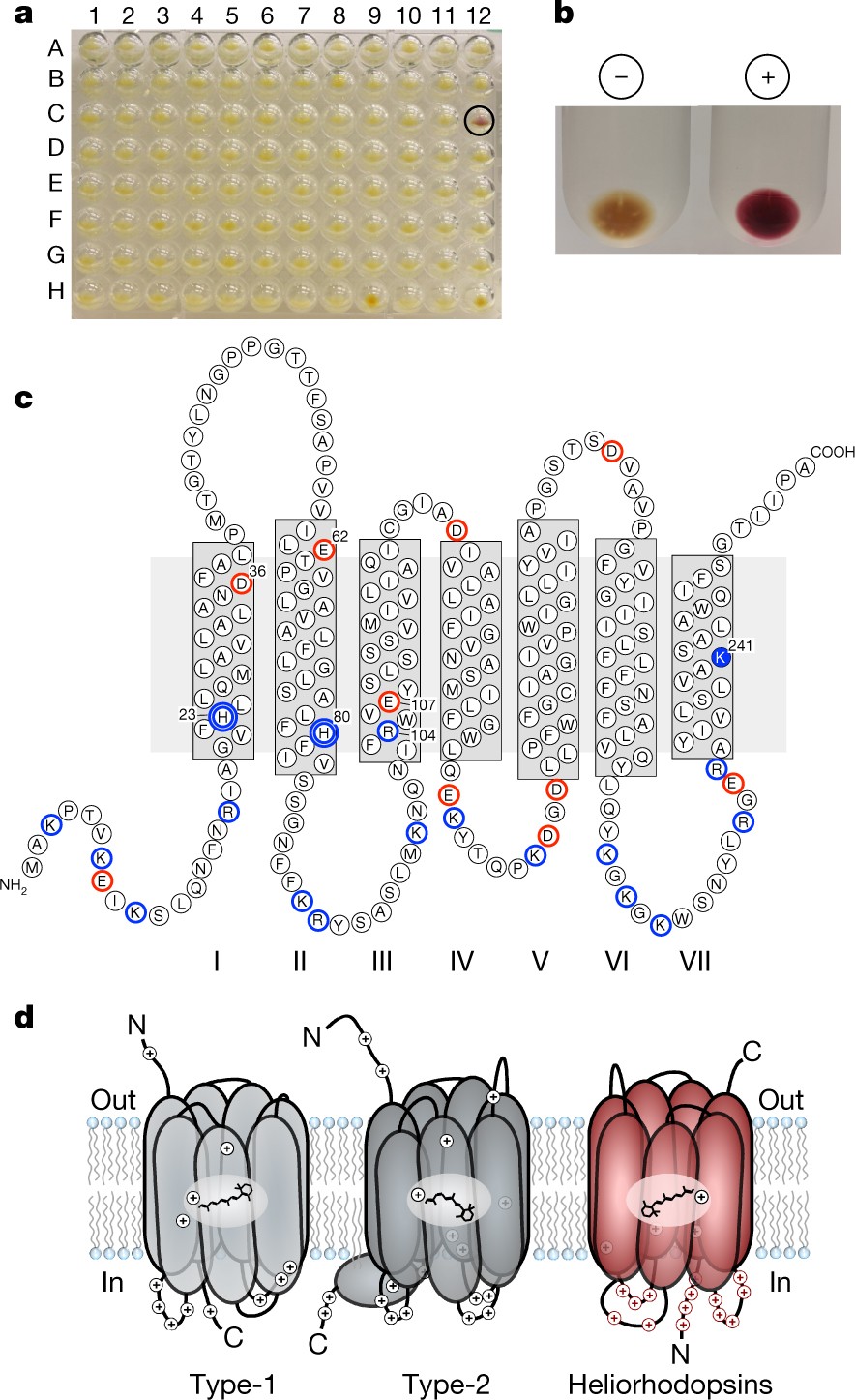
The second library screened was from a freshwater lake (Lake Kinneret / the Sea of Galilee). This time we were also scoring positive clones by color. One 96-well-plate, #48, gave a single clone with a distinct deep purple color (see insight in the figure below). This color was observed only when retinal was added, so we were expecting a known microbial rhodopsin. To our surprise, and actually disbelief at first, no rhodopsin gene was observed in the sequence obtained from this clone (a 32 kb insert). As we were flying blind, Tn5 mutagenesis screening was used to detect the gene responsible for the purple color. This gene turned out to code for a 7 trans-membrane helices (TM) protein with a lysine in the 7th TM and inverted membrane topology compared to any known rhodopsins. In addition, this protein had less than 15% identity to known microbial rhodopsins, the reason it was not detected in BLAST searches as a rhodopsin. The detection of this new protein enabled us to find a whole portfolio of novel rhodopsins, heliorhodopsins, in archaea, bacteria, algae and even in viruses (Pushkarev, Inoue, et al. 2018).
We are now in the process of sequencing and analyzing a dozen more colored clones from the same library.

The paper in Nature is here: https://go.nature.com/2K7rVgC








Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Definitely worthy of 'smoke on the water'!