The paper in Communications Biology is here: go.nature.com/2mvaPMb
The ideas of synthetic biology to engineer biology for making devices and “biological machines” is really thought-provoking. One way we have seen this is through iGEM, where students are absolutely thrilled about the ideas of synthetic biology. Can we really engineer biological systems the same way electronic or mechanic devices are made? While working on ideas of how to make microbes produce patterns and structures we realised that a central concept is to distribute different tasks between populations of cells. In a sense we were thinking of how to engineer microbial populations to work together in a “multicellular” way.

One of the inspirations for this work was the different patterns and structures in biological materials. Here is an electron micrograph of calcite crystals packed in the shell of a blue mussel. The width of the image is 100 micrometer (photo credit Pezhman Mohammadi).
We are starting to understand the importance of microbial languages and communication more and more. Great examples of large and complex microbial communities are the gut microbiome and the plant rhizosphere that play an important role in health and defense against pathogens.
Microbes are incredibly diverse and have specialised to function in nearly any condition on earth. In a perfect scenario, we would be able to select microbes based on their natural specialisation, and then combine/complement their functions in artificially constructed microbial communities. The range of possibilities, such as making materials, the conversion of waste streams, and the production of chemicals, would be almost unlimited. In reality however, we know that microbial interactions are inherently very complex and hard to predict. There are some obvious difficulties we face when trying to create artificial microbial communities. Differences in growth rate, differences in expression levels, different nutrient requirements, randomised physical interactions and mixing of cells, make it challenging to control and study even two cellular populations.

Here is Chris, one of the co-authors, working at the microscope.
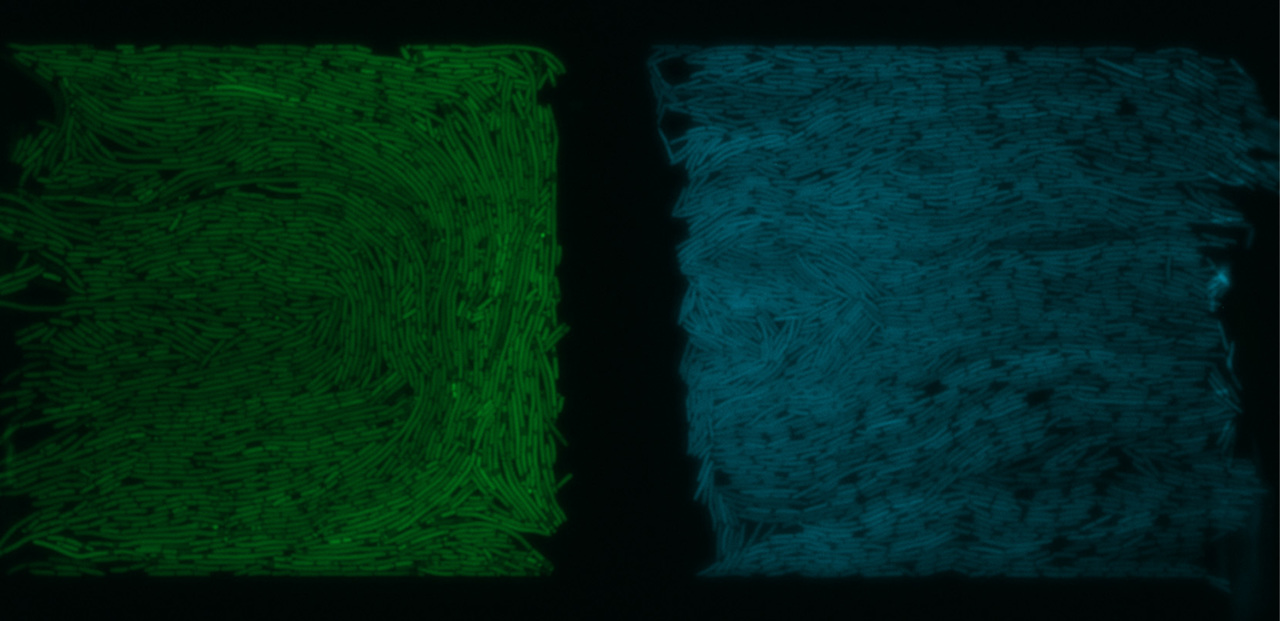
In our paper, we describe a microfluidic device in which we have complete control over two bacterial populations and their interaction. In this, the populations are physically separated with a thin filter allowing the chemical communication between them. A sender population controls the expression of several genes in a receiver population. We first showed a stable “on” interaction over long periods, to demonstrate the stability of communication. We then looked at oscillations, in which the sender population continually switched from “on” to “off” state and saw that the receiver followed the behaviour perfectly without significant delay.

Trapped and communicating in a microfluidic device. Two populations of E.coli separated by a membrane are able to communicate but not mix (photo credit Chris Jonkergouw and Ekaterina Osmekhina)
Achieving both stability in the populations and control over their interaction, this method provides a first step towards the programming of complex microbial communities. In the long run we might be able to build networks like in computers. Computers allow us to do a huge variety of things, although the electronic parts that make it function, consist mainly of a large number of standardised logic gates that use a simple rule to convert inputs into outputs, both consisting of “0” or “1”. If we can find a way to standardise the parts (logic gates in computers, microbial populations in our case), and then define a common rule for their function and interaction (“0” and “1” in computers, expression “on” and “off” in our case), we can, like computers, start to scale the function and range of applications of microbial communities. But, in order to functionalise a community, we are first need to control, stabilise and standardise the individual populations and also their interactions.
Here is a link to our paper ttps://rdcu.be/3fzk
This post was written together with Chris Jonkergouw and Ekaterina Osmekhina




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in