A recent paper by Adam Martiny critically examined the old saw that "only 1% of microbes are culturable" [1]. In this paper, Martiny pointed out that this statement can mean one of three different things: either that 1% of microbes in an environment are culturable, that 1% of taxa are culturable, or that 1% of cells in a community grow on agar plates under standard conditions. What's worse, most authors don't clarify what, precisely they mean by this statement, and it isn't even clear who deserves credit for this idea, or what data initially stood behind it. Many microbiologists (including myself, for something like 15 years) simply assumed this was true.
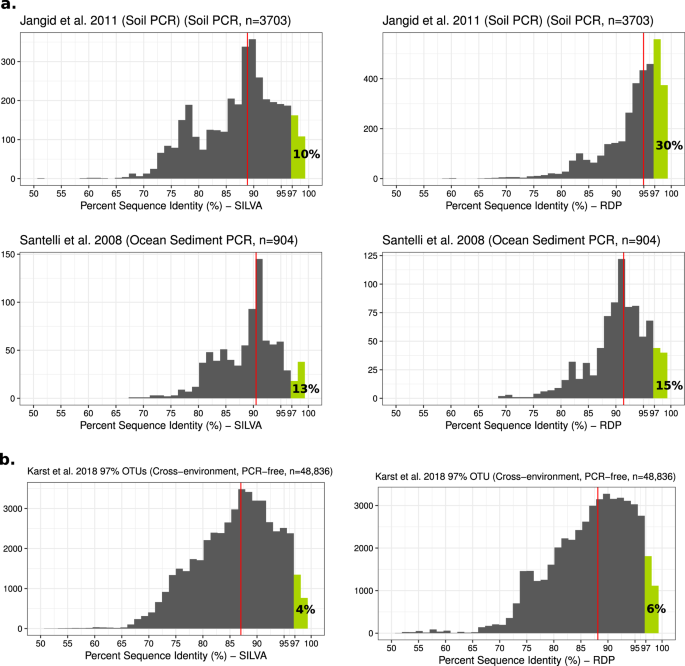
I was pleased to see Dr. Martiny so clearly lay out the problems with the paradigm of 1% culturability. I was surprised, however, to see his conclusion that something like half of both cells and taxa are closely related to cultured taxa, albeit with wide variation among environments and studies. Karen Lloyd and I had published a paper that came to the opposite conclusion about a month after Dr. Martiny submitted his paper [2]. In that work, we drew 16S rRNA sequences from more than 5000 publicly-available studies, including 16S amplicon libraries, metagenomes, and metatranscriptomes. By identifying the nearest cultured relative of each of those sequences, we estimated that 81% of bacterial and archaeal cells on Earth belong to uncultured genera, and 25% belong to uncultured phyla.

These conflicting findings may have just gone unnoticed if Cameron Thrash (@jcamthrash) had not tweeted a link to the Martiny paper, which sparked a wide-ranging discussion. Within an hour and a half, Alex Crits-Christoph (@acritschristoph) analyzed a large 16S data set from soils to show that very few 16S sequences were closely related to cultures. Paul Carini (@paulcarini) pointed out that Martiny's use of PCR-amplified sequences seriously biases results towards cultured species, since 'universal' primers are well known to miss large groups of deeply-branching phyla. Paul also pointed out that he, Kristen DeAngelis (@kristenobacter), and Cameron Thrash are working hard to develop innovative new ways to culture microbes, and Alex pointed out that Noah Fierer (@noahfierer) had written a blog post raising many of the same points that we had. None of our group knew all the other members personally before we wrote the paper, but we formed like Voltron, moved the discussion offline, and collaboratively wrote a manuscript in about two weeks, which has now been published. In the end, while we disagree with Martiny about the fraction of Earth’s microbes and microbial taxa that have been cultured, we agree that advances in culturing techniques and intensified efforts will lead to ‘unculturable’ microbes being grown in culture.
Social media exerts a deep influence on our society, often not for the better. Our new paper, however, is a clear result of online communication that would not have been likely to happen IRL (that means “in real life”).
References
Header image credit: Kristen M. DeAngelis.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in