In 2013, Hauke Smidt, Detmer Sipkema, and me initiated a research line: “Tapping into the natural halogen cycle”. This was a complementary team considering the interest of Hauke and me on microbial dehalogenation, and Detmer on microbial halogenation. We presented the slide below in December 2014 at the strategic meeting of the Laboratory of Microbiology at Wageningen University & Research: our plans for upcoming years.

Our intention was to better understand microbes in natural pristine environments that add halogens to natural organic matter and those that remove halogens, and if there were any interconnections between halogenation and dehalogenation. Together with other like-minded colleagues from the Laboratory of Microbiology and beyond, we later discussed our thoughts and visions in two comprehensive review papers published in Environmental Microbiology and Frontiers in Microbiology.
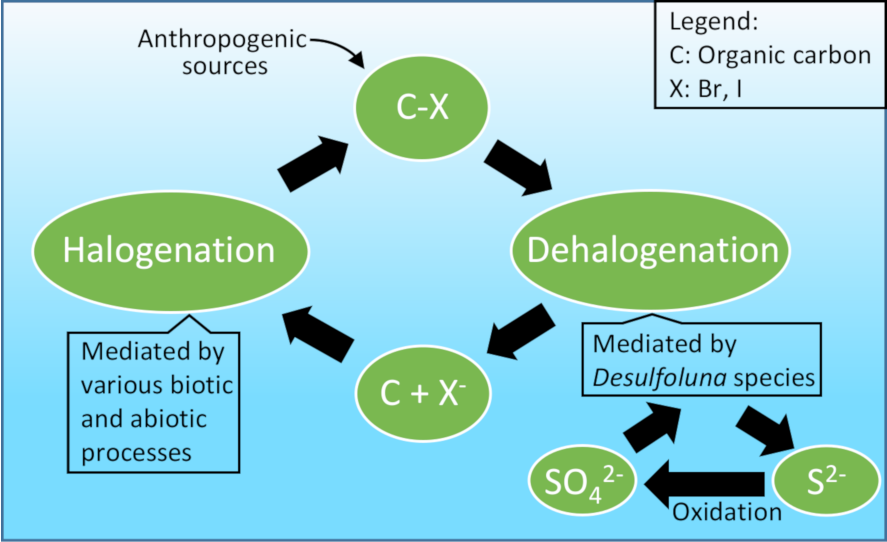
Owing to the toxicity of many halogenated organic compounds (organohalogens), halogenation is employed by (micro)organisms in many cases as a chemical defense mechanism. Not surprisingly, some organohalogens like chloramphenicol and vancomycin have found important pharmaceutical applications. In turn, microbes can remove halogens from organohalogens. A special group of anaerobic microbes known as organohalide-respiring bacteria (OHRB) use organohalogens as their electron acceptor and grow while removing the halogen substitutes. They essentially breathe organohalogens as we breathe the air.
The OHRB were discovered in a quest for bioremediation of sites contaminated with manmade organohalogens, some of which are among the most toxic compounds on Earth. However, presence of OHRB in pristine environments is not surprising considering thousands of known natural organohalogens (some produced at the scale of gigagrams per year) and their ancient history on Earth that predates the industrial era.
With this in mind, Detmer and his PhD student Johanna Gutleben started collecting marine sediments in the vicinity of sponges that are known sources of organohalogens, and hence likely to harbor OHRB. Rather than marine sediments, the first batch of samples turned out to be intertidal shore sands from Spain, because the sponge sampling site was too rocky to collect any sediment.

OHRB are usually strict anaerobes that thrive under organic-rich conditions, but the sands we got were exposed to the air and did not seem very rich in organic matter either. This was the period when Peng Peng joined us as a PhD student who was very eager to start his research. I didn’t think we would find any OHRB in those course sand samples but I remember Hauke saying “Let Peng get his hands dirty. You never know what comes out.” And he was right! The hard work of Peng in the past five years payed off, and he could isolate an organohalide-respiring Desulfoluna strain which resembled in many ways the bacterium previously isolated from sponges by the group of Max Häggblom. This has led to collaborations with the group of Max and other research groups, and comprehensive insights into the physiology, genetics and biochemistry of marine OHRB which is now published in ISME J. In another parallel research line, we found reductive dehalogenation potential in sediments obtained from pristine hypersaline lakes in Australia.
Anaerobic dehalogenation in pristine environments is gaining more importance, and a special session has been devoted to this research avenue in the upcoming dehalogenation conference, Dehalocon 2020. Putative reductive dehalogenase encoding genes are increasingly being found in the genomes of both cultured and uncultured bacteria, archaea, members of candidate phyla radiation (CPR), and even human gut microbiota. Proving that these putative genes are indeed involved in reductive dehalogenation will be the next hallmark discovery, and a key for guiding cultivation of microbial dark matter carrying these genes.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in