Evolution has brought us the remarkable diversity of life and is a constant source of inspiration in biotechnology. Unfortunately, the very same adaptive mechanisms threaten human health when pathogenic bacteria evolve to resist the antibiotics that doctors rely on for treating infections such as UTIs, pneumonia or even worse.
Antibiotic resistance can be either invented or borrowed
Fundamentally, bacteria can evolve resistance through two mechanisms: the alteration of existing genetic material in their genomes (vertical evolution) or via the acquisition of foreign genetic material (horizontal gene transfer or HGT in short).

Fig. 1. Antibiotic resistance evolution. Bacteria may mutate their genome or acquire novel genetic material to become resistant. However, little is known about the interactions between these two modes of resistance when present in the same cell.
My main focus has always been on the horizontal aspect of resistance evolution, due to its critical involvement of resistance acquisition in some of our most important pathogens. However, mutational changes can, without external genetic contributions, render bacteria resistant to antibiotics, and arise readily in lab evolution experiments, but seems to be less involved in clinical resistance for most bacteria.
These two modes of resistance are almost exclusively studied in isolation; however, there is no reason to believe that they do not occur together in nature. In fact we do observe them together in sequenced genomes (Fig. 2)
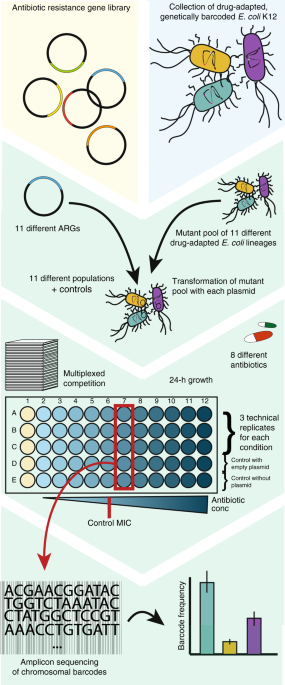
With previous work performed on both horizontally acquired and mutational resistance in our lab, we set out to investigate how these two modes of resistance would potentially interact when present together in the same bacterial cell. From these studies, we had a diverse collection of well characterised strains being resistant through either mutations or resistance genes. This was a great starting point and a good opportunity to do a fun and collaborative project that combined the expertise within our group.
We simply imagined a scenario where a resistance gene is transferred to a mutant strain – what kind of interaction would there be? Would both modes of resistance persist, or would one be preferred?
After assaying more than a hundred gene-mutant combinations in different antibiotics, we saw that some resistance patterns would deviate from that of the individual resistance determinants. Notably, in most cases, there would be no benefit of having resistance mutations if a resistance gene of similar resistance spectrum was present; unless the bacteria were challenged with very high antibiotic concentrations. In addition, we observed that a widespread tetracycline resistance gene (tetA) was able to cancel out the effect of mutations conferring resistance to the unrelated aminoglycoside antibiotics, and we show that this is likely to be a general effect of efflux mediated resistance.
We included a small bioinformatic analysis in the paper, showing that the nuo mutations leading to aminoglycoside tolerance and the tetA tetracycline resistance gene was not observed together in clinical E. coli genomes. If we expand the analysis to include all complete E. coli genomes available from NCBI’s refseq database, we can speculate a little bit further on the general implications of our experimental observations.

Fig. 2. Co-occurrence of ARGs and mutants in sequenced E. coli genomes. (A) Bars show the observed relative to the expected proportion of gene-gene (GG), gene-mutation (GM) or mutation-mutation (MM) co-occurrence events in ~13.500 sequenced E. coli genomes. (B) The proportion of negative (less frequent co-occurrence than would be expected by chance), positive (more frequent linkage than would be expected by chance) and random co-occurrence events within each interaction category.
As would be expected, due to co-localization on mobile elements such as plasmids, antibiotic resistance genes occur much more frequently together than do mutations; however, co-occurrence within both categories is more frequent than would be expected by chance (2.7 and 1.06; χ²-test, P < 0.001). In contrast, the co-occurrence of ARGs and mutations is observed less frequently than would be expected (0.94, χ²-test, P < 0.001) and there is a higher incidence of negative interactions between these. The low incidence of positive co-occurrence between the two modes of resistance supports our experimental findings where orthogonality, redundancy and negative interactions were mostly observed.
In summary, our study indicates that bacteria exposed to antibiotic concentrations used in the clinic (below the clinical breakpoint) resistance genes tend to be favoured over resistance mutations, and that resistance mechanisms to unrelated antibiotic classes may also suppress each other. This complexity is important to consider when studying resistance evolution and may help explain why resistance mutations are rare when transferable resistance genes are readily available.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in