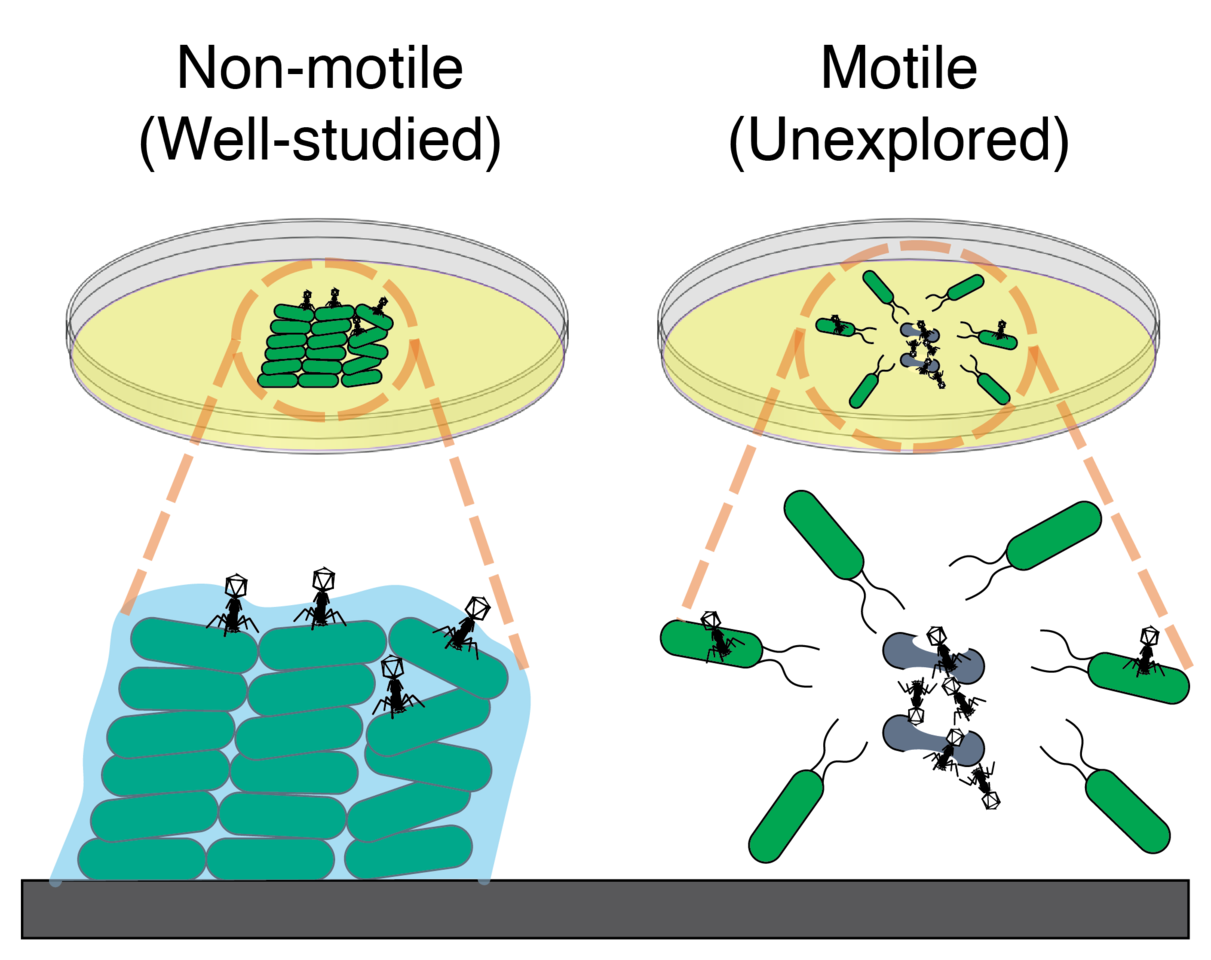
The idea behind this project came from a casual meeting between three coauthors: Seppe Kuehn who studied how chemotactic E. coli evolve faster migration (eLife, 2017) and Kim Sneppen and Sergei Maslov who are long-time collaborators with a keen interest in phage. A question naturally emerged: how do phages infect chemotactic bacteria? In the past, studies of phage infection in space focused on what happens in the bacterial lawns or biofilms (Fig. 1 A, left). Those studies shed light on how phage attack non-motile bacteria. Surprisingly, how phage infects and coexists with moving bacterial populations was largely unexplored. To study this question, Derek Ping, an undergraduate student from Seppe’s lab, performed experiments by inoculating the chemotactic E. coli cells together with their phage P1vir at the center of an agar plate with rich medium (Fig. 1A, right).

When Derek initiated such an experiment and then waited 19 hours he observed a striking pattern like the one shown in Fig. 1B. The outermost bright rings are dense bacterial populations which are migrating at about a half a centimeter per hour (the density billions of cells per cm3). These outer bands are well studied (Adler, Science 1966) and arise from growth and chemotaxis. However, at the center of the colony, Derek observed a darkened area, about 6cm in diameter, which he showed resulted from the collapse of the bacterial population due to phage lysis. Further, at the center of the colony, he observed a dense region due to the rise of resistant bacteria.
We sought to understand how the phage could create the large central region of the colony where the bacterial population had collapsed. Existing theories based on studies with non-motile bacteria showed that phage could not move over such large distances (centimeters) in such short periods of time (hours) without being actively transported. Therefore, we speculated that the phages travel along with migrating bacteria either during the latent period of infection or while attached to the cell prior to injection. To test this hypothesis, we built a mathematical model that included the ability of phage to “hitchhike” with migrating bacteria. The model confirmed our hypothesis.
We then set out to study phage hitchhiking with chemotaxing bacteria depends on phage infectivity and population size. Further experiments revealed two regimes: one at low infectivity or initial phage populations where the migrating bacterial population “escapes” from the hitchhiking phages. In contrast, at high infectivity or initial phage density the virus collapse the bacterial population and bacteria have to evolve resistance to survive (Fig. 1C).
Overall, our results show that phages can hitchhike with migrating bacterial populations. In this scenario, either infectivity or initial viral loads are high enough to collapse the bacterial population, or they are low enough such that the bacteria can escape. Our work opens a window into the understanding the eco-evolutionary struggle between phage and their bacterial hosts in the context where the predator (virus) can hitch a ride with the prey (bacteria).





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in