To enjoy the full story, check out our Nature Microbiology paper: “Pervasive transmission of a carbapenem resistance plasmid in the gut microbiota of hospitalized patients”.
“A Warm Welcome”
Since I was a University student I have been captivated by mobile genetic elements and particularly by plasmids and their dissemination. In fact, these elements are the main actors in the spread of antibiotic resistance in bacteria, which is one of the most concerning health challenges worldwide. During my PhD, I started studying how these plasmids spread, and I wondered if we would be able to track them across hospital settings. Four years ago, when I joined the Plasmid Biology and Evolution (PBE) Lab lead by Alvaro San Millan, I had the chance to answer this question.
“Inside Information”
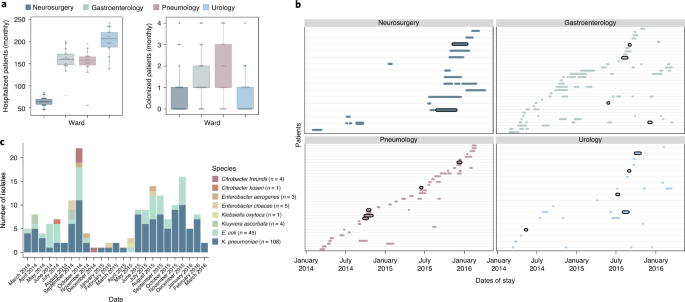
Let’s start from the very beginning. The R-GNOSIS project included the isolates and metadata recovered from more than 9,000 patients sampled on a weekly basis for two years in the Ramon y Cajal University Hospital in Madrid. This collection was an ideal resource to track and understand plasmid dissemination in a hospital setting. To reach this goal we had to collaborate with different departments within the hospital and overcome one of the main challenges in the scientific community: to integrate and forge good connections between different disciplines. Our collaboration with the Department of Preventive Medicine was crucial, providing us with one of the cornerstones for our study, the metadata from all the patients admitted to the hospital which they collected side by side with the Microbiology Department for two years.

Between-patient transfer of plasmid-carrying enterobacteria in the hospital. Each row represents an individual patient admitted to the ward and the arrows represent transmission events.
“Not At Home”
After obtaining all the metadata from the patients I needed to find a way to analyse the database to decipher how the plasmid was disseminating in the hospital, so we decided to contact Prof. Ben Cooper, a world-class expert on using mathematical modelling and statistical techniques to understand infectious diseases dynamics. Prof. Cooper was based at that time at the Mahidol- Oxford Tropical Medicine Research Unit in the city of Bangkok, so I decided to travel there to start my quest towards discovering epidemiological models and, moreover, towards discovering Thailand.
Once there, a new challenge emerged. To dive into this new discipline, I needed to adapt my mental framework. The first and most important skill to learn was to understand the importance of the database itself. I needed to merge and curate the databases obtained from four different wards and collected by different staff over two years. Carrying out this effort allowed me to ask the right questions. Alongside the extremely friendly and welcoming Cooper’s team and combining mechanistic models with the epidemiological and genomic data we disentangled the relative importance of the between-patient (i.e., patient to patient) and within-patient (i.e., spread in the gut of each patient) plasmid transfer in the hospital, highlighting the extremely high frequency of within-patient plasmid spread between different bacteria in the gut of hospitalised patients.

From left to right, Ricardo León-Sampedro, Mo Yin, Célia Souque and Thomas Crellen (co-author in this work) visiting a temple during a trip to Myanmar. For those of you wondering, the cream on RLS’s face is Myanmar's thanaka cream, for sun protection.
“The Return Journey”
Once we demonstrated the importance of the within-patient plasmid dissemination, we wanted to find specific events that would illustrate this process. To achieve this goal, I found four examples of genetic fingerprints in which the same plasmid variant was present in different bacterial species in the same patient, confirming that these were indeed plasmid transfer events between different bacteria that occurred within the gut of a patient.
What have we learned from our results? We showed that the frequent between-patient transfer of plasmid-carrying enterobacteria is driven by high-risk bacterial clones. Once patients were colonised, there was a pervasive conjugative transfer of the plasmid in the gut of patients, contributing to its long-term maintenance in the gut microbiota. With these results, we provide new insights for the development of new intervention strategies able to control the in-hospital spread of antibiotic resistance.

Within-patient plasmid transfer. Once a patient is colonised by plasmid-carrying enterobacteria, we track rare (traceable) plasmid variants spreading among different bacterial species.
“The Last Stage”
This work was possible through the efforts of many different people. In the PBE-lab we worked as a real team, and all the discussions, the sharing of new ideas, or solving the problems together strengthened not only the results but also the quality of our science. This way of working facilitates the interaction with other teams, therefore making multidisciplinary works like this one possible.
I hope you enjoy the paper as much as we enjoyed the journey to get here. This study opened up new questions that we will be answering in our upcoming works.
Stay tuned!







Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in