Viruses are tiny, we all know that from our textbooks. Well, actually not all of them. Over the last fifteen years weird viruses with huge particles and genomes were discovered. These so-called “giant viruses” have genomes that for some of them are larger than the ones of several cellular organisms. They also have viral particles even bigger than the tiniest eukaryotes.
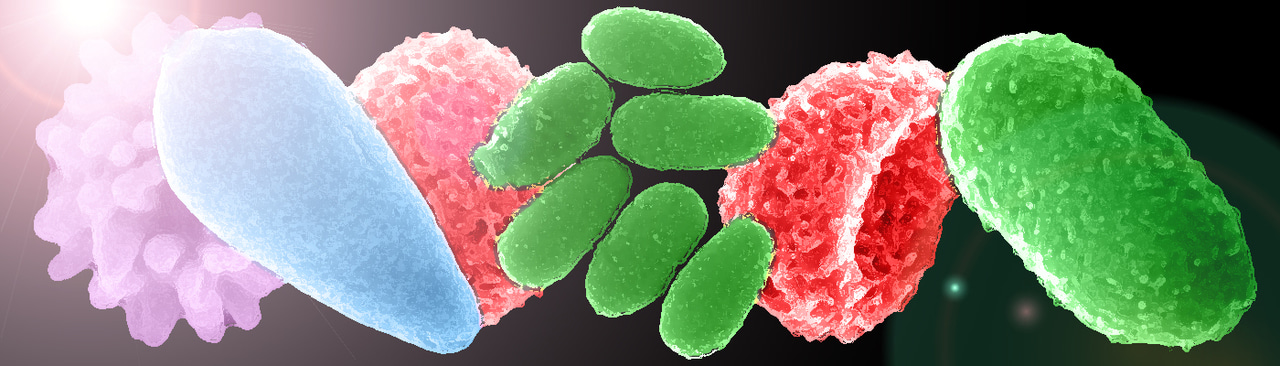
We, and other groups, have contributed to the discovery of new families of giant viruses in recent years. They mostly infect amoebas but also a wide range of eukaryotes. The hunting bag is now filled with a variety of giant viruses with different shapes, genomes, gene contents, replication strategies and so on. The apparent diversity of giant viruses, exemplified by their various morphologies (see Figure), is now well established thanks to the deluge of metagenomic data that has emerged in recent months on this subject. So yes, indeed, they are extremely diverse and found in various environments.

SEM images of viral particles of Mollivirus sibericum (red), Mimivirus (purple), Pithovirus sibericum (green) and Pandoravirus salinus (blue). Melbourvirus particles are also shown (orange) using a cyro-EM image (right image) and 3D reconstruction (left image). © Lionel Bertaux – Chantal Abergel – Matthieu Legendre
Genomics and metagenomics data now help to get a clearer picture of the genes and biological functions encoded by giant viruses. We also know more on how their genes evolve. But we are still missing one piece: their epigenomes. Indeed, besides the genes DNA also provides other types of information called “epigenetics” by the biochemical modification of the histones (the proteins that pack the DNA) or the nucleotides themselves. In cellular organisms, modifications such as the methylation of the DNA play a critical role in gene regulation, DNA replication or cell cycle regulation. So what about giant viruses? Do they also bear this type of epigenetic information? We have recently addressed this question and published our investigations in Nature communications.
Before I expose our results let me first briefly explain how we spotted the modified nucleotides in giant viruses’ genomes. We actually benefitted from the recent advances in long-read sequencing technologies. These techniques, named SMRT (for Single-Molecule Real-Time sequencing), not only record the sequence itself but also the time it takes for the polymerase to incorporate nucleotides. Since that duration is different whether the template DNA is modified or not, it gives a way to track methylated regions of the genome at a single base-pair resolution. Some of the giant viruses we recently discovered were sequenced using this technique. We thus gathered these datasets and resequenced other giant viruses as well as newly isolated ones with the same approach. All together, we obtained epigenomic data covering a large spectrum of today’s known giant viruses’ families.
What have we learned from these data? First that DNA methylation is widespread in giant viruses. It affects almost ¾ of the giant viruses we analyzed. At the same time their genomes are not uniformly modified, some are only slightly modified while others contain much more methylated DNA. We also discovered that the enzymes responsible for this methylation, the methyltransferases (MTases), are encoded by the viruses themselves. So, where did they get those genes? One might expect that they got them from their hosts. After all that’s what viruses are supposed to do, steal genes from their hosts. But in this case they got them from the bacterial world. It is important to know that amoebas (the host of all the giant viruses we studied in this work) are associated with bacteria because they feed on them. Thus, giant viruses are continuously confronting them. Our evolutionary analyses show that the MTases’ journey is even more complex than previously imagined. After robbing bacteria, the giant viruses passed the MTases to their hosts and also exchanged them with each other. Most of the time the MTases are conserved within the viral genomes, suggesting that they increase virus fitness.
What could be the function of DNA methylation in giant viruses then? We believe that it could be involved in virus-virus interactions. For some of the viruses, the MTases are encoded next to endonucleases that cleave the DNA at specific target sites, identical to the ones recognized by the MTases. This forms the so-called “Restriction-Modification” (R-M) systems. These are very common in bacteria where they allow them to discriminate their own methylated DNA from the unmethylated one of invading bacteriophages. The bacteria can then fight against phages by cleaving their DNA. In the case of giant viruses this could be a way to fight against competitors when they co-infect the same host. Indeed life of a virus is not peaceful, even for the big ones. They constantly have to fight each other to replicate within the host. This is particularly true for viruses infecting amoebas, which are prone to infection by a large variety of viruses and intracellular bacteria.
If R-M systems could be a weapon of choice, most of the giant viruses’ MTases are not associated with endonucleases, they are “orphans”. In that case we believe that it provides the virus a nice self-defense response against competitors harboring complete R-M systems. Methylating their DNA shield them against the other viruses and bacterial endonucleases, making them immune to their attacks.
The analysis of giant viruses’ epigenomes is now technically feasible under the metagenomics framework. We expect that such data will be published in the future and hope that it will provide clues as to how it shapes their interactions with each other and with their hosts.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in